Game on! - learning to fight real diseases in a virtual world
Picture a university laboratory where molecular-level research is carried out into biological processes. The laboratories are a hive of activity among samples, test tubes and cell cultures. One laboratory stands out from the others: here too work is performed on molecules and chemical reactions, but there is not a single flask or pipette to be seen. Here there are only computers, and a group of researchers glued to the screens.
These are the members of the Molecular Modelling Group at the 'Severo Ochoa' Centre for Molecular Biology at the Autonomous University of Madrid. Headed by Dr Paulino Gómez-Puertas, the team is made up of David García, Fernando Martín, Dr Jan-Jaap Wesselink, Dr Jesús Mendieta and Dr Eduardo López-Viñas.
The few computers that are on view are not really the ones doing the work. What you can see are just terminals connected to the real calculating machines, housed in a specially acclimatized room at the other end of the campus, a kilometre away. Inside those machines the team is manufacturing an unreal world that is intended to be an image of the actual physical world we live in. They produce virtual nucleic acids and proteins, and provide them with movement thanks to enormously complicated calculations in which the forces that govern the interactions between atoms within the molecules are analysed and simulated. In this overwhelmingly tiny virtual world, distances are measured in millionths of a centimetre while the atoms attract each other, collide and separate at terrifying speeds, over periods that are measured in trillionths of a second. And thanks to the movement of the atoms that make them up, the proteins also come to life. They vibrate and twist; they join up to other proteins; and they take part in chemical reactions. All this is in an effort to simulate, in a universe that does not exist, how all these things really do occur in the cells of a parallel universe?in other words our universe, reality.
From all this information, immersed in their simulated world, Eduardo and David are building an automatic drug design system. To do this they use 3D images of the space occupied by the proteins and systematically try to fit millions of different chemical compounds into the hollows that appear in the surfaces. This is no mean feat; not only do they have to look for the right shape and size, they also have to weigh things up on the atomic scale. At this level electrostatic charges are a nightmare, and there is no way to keep electrically charged molecules still. Despite the difficulties, they are making good progress, particularly in the design of drugs that target proteins involved in diabetes and obesity.
Jan-Jaap does not work with proteins, but with the incredibly long nucleic acid sequences that carry genetic information and make up our DNA. These are chains of hundreds of millions of small molecules known by the letters A, G, C and T, which he tries to make sense of. Contained within these sequences are the blueprints of the virtual universe, but you have to know how to interpret the plans in order to build correctly without making mistakes, just as in the real world. The same is true if you want to interpret the errors that can bring about illnesses such as cancer. Jan-Jaap sets about this task using the daunting amount of new information that 'next-generation sequencing' techniques generate, as they revolutionize the world of DNA.
Jesús and Fernando work on the detailed simulation of the enzymatic reactions that take place inside proteins. In their virtual world they want to be able to mimic the smallest detail of the steps that transform one chemical compound into another. They hope to understand how proteins manage to facilitate such violent reactions inside something as delicate as a living cell. The space they work in is populated by tiny molecules packed with energy, of metallic atoms and of water molecules that interact with all the others and take part in the reactions. It is a long slow process, but it is starting to yield results in simulations of certain bacterial proteins that are used as targets in the design of a new generation of antibiotics. And this is just the start. The dream of all these researchers is to be able to build an exact working replica of a real cell in their virtual world, with all the atoms interacting with each other in the same way as they do in the physical universe. When they manage to do that, it will no longer be necessary to carry out experiments in test tubes and petri dishes; a good internet connection will be all you need to test the effects of millions of compounds on a protein or on a metabolic pathway, and to choose the best to produce as a drug. There is still a long way to go in terms of time and effort before they realize this dream, but the Molecular Modelling Group is taking the first steps.
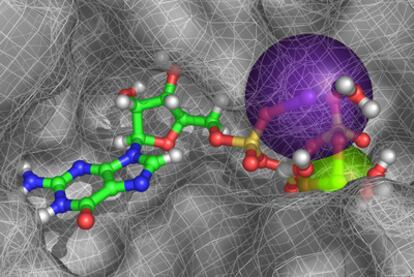
Figure 1. In virtual space, a guanosine triphosphate molecule nestles into a perfectly-shaped hollow in the surface of a bacterial protein, while it interacts with a magnesium atom (green sphere), a potassium atom (purple sphere) and four water molecules (red and grey structures, on the right).
Paulino Gomez-Puertas, Universidad Autonoma de Madrid.- www.atomiumculture.eu