Exploiting Biomedical Information through Literature and Ontologies
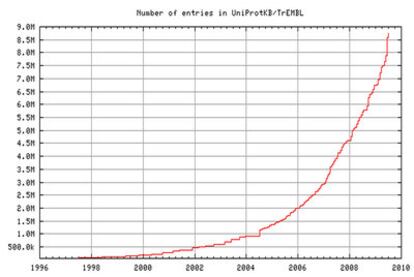
However, identifying what exactly a given protein does is a difficult task not only because of the sheer number but also because the function of each protein is normally regulated by other proteins. Although the sequence in which amino acids - the building blocks of proteins - are arranged is known in the case of 9 million proteins, only 400 000 are known in terms of their function. As the techniques of determining the exact sequence of amino acids in a protein - its unique fingerprint - become more powerful, the number of known proteins keeps rising exponentially (Figure 1)?which traditional manual methods to determine the function of a protein can never catch up.
It is to meet this challenge that automatic tools have been created to predict the functions from what we already know about the functions of similar proteins. However, this approach is complicated by two observations, namely that some proteins are unique - no existing protein is quite similar to them - and that a similar sequence does not always mean a similar function. Lastly, no matter how the function of a protein has been arrived at, researchers still need to verify that function before they can use that information?and the process of verification is complex if no clear and sound evidence is provided.
In identifying the functions of a protein from the proteins similar to it or even in proposing that a given protein has novel functions, researchers often rely on published biomedical literature, a hard and time-consuming task even for experts and made harder by the sheer volume of published literature. For example, PubMed, a commonly used database of documents in the life sciences, currently comprises over 16 million records. In 2008 alone, 671 904 documents were added to the database. Even if a researcher were to read all of them, reading 10 documents a day, the task would take more than 180 years? Thus, we certainly need more efficient strategies to exploit the literature.
One such strategy is to use automatic text-mining systems, and although they have been used successfully to identify the functions of new proteins, most of such systems rely on manually coded knowledge in the form of rules and examples provided by experts. To overcome this limitation, I decided to explore whether this process too can be automated, that is to find out ways of constructing the relevant rules and choosing appropriate examples automatically by using ontological information instead of relying on experts to provide the rules and examples.
I implemented this strategy in GOAnnotator, a web-based tool that assists in identifying and verifying protein functions from biomedical literature. The user gives a protein, and GOAnnotator not only finds relevant texts that point to the probable functions of that protein by automatic extrapolation but also refines the predictions based on what those texts say. GOAnnotator relies on the extrapolated protein's annotations that describe protein functions using the controlled vocabulary of Gene Ontology, thereby avoiding the sometimes ambiguous descriptions which can result from using free text (uncontrolled vocabulary). GOAnnotator uses domain knowledge encoded in the ontology to mine the text of biomedical documents automatically to find those that supply suitable evidence for predicting protein functions.
Compared to the traditional approaches, GOAnnotator is more effective. A team of curators volunteered to use GOAnnotator to verify the functions of 66 proteins and found that GOAnnotator was able to extract relevant texts from the biomedical literature with a precision of 93%. This high precision provides substantial evidence that automating the exploitation of domain knowledge from ontologies is a valid alternative to using domain knowledge explicitly created by experts.
GOAnnotator has other advantages as well. GOAnnotator requires minimal human intervention, since it avoids the complexities of creating rules and patterns covering all possible cases or generating example sets that are too specific to be extended to new domains. Secondly, GOAnnotator covers domain knowledge more extensively than human operators can. Lastly, GOAnnotator's results can be always up to date because it tracks public data sources automatically for updates as they evolve.
GOAnnotator may appear a niche application but the principle behind it has immense potential. The explosion of knowledge in the life sciences, and the pace at which it continues to develop, make the tasks of digitizing, storing, and analysing that knowledge appear insuperable. Knowledge is power but only when it is harnessed efficiently?and GOAnnotator shows a way to do just that.
GOAnnotator and other tools following the same approach are freely available at.